- Drag the Table button from the Object palette:
- This will place a 3 x 3 table on the page.
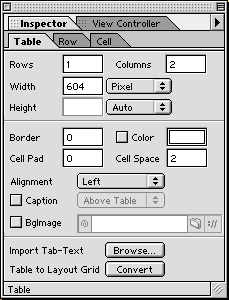
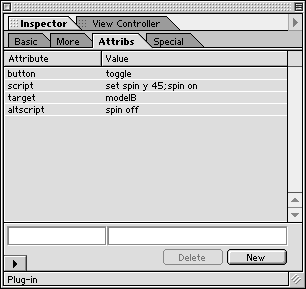
- To select a cell within the table, click on the bottom border of a cell. To select the entire table, click on the upper border of the table. The Inspector palette will change depending on what is selected. The Inspector palette can be used to set the number of rows and columns in the table. It can also be used the set the width of the borders and the padding between the borders and the cell contents.
- If the border is set to 0, then the table grid will not be seen: