Overview
Benzylpenicillin (penicillin G) and phenoxymethylpenicillin (penicillin V) are produced by fermentation and are the foundation for a range of synthetic antibiotics. Penicillin acylases, which are found in microorganisms such as bacteria, yeast, and fungi, are used to catalyze the hydrolysis of natural penicillin’s. It is also used in the hydrolysis of cephalosporin’s, which yield another group of common antibiotics. The amide link shown in the figure below may be hydrolyzed normally but the conditions for this reaction to occur are difficult to obtain. The hydrolysis instead is simply achieved by the use of penicillin acylase. The use of this enzyme is one of the earliest successful processes involving immobilized enzymes and may be reused over 100 times. The penicillin G acylase can be used in the reverse of the reaction as shown in the schematics below.
In an article by McVey on the structure of penicillin acylase, insight is given about the catalytic mechanism and structure of the enzyme. Not much is known about the physiological role, which they speculate to be involved in the assimilation of aromatic compounds. There is however much known about the substrate specificity. The acylase enzyme hydrolyzes a number of amides, which are generalized by the formula below:
R-CO-NHR’
The specificity of the substrate is determined mainly by the acyl (R) group, which means the enzyme demonstrates an attraction for hydrophobic groups. The R’ group however can vary widely and the leaving group of the substrate will have minimal to no effect on the hydrolysis rate.
Structure and Active Site
In the E.coli penicillin acylase, there are two monomer chains (A and B), which consist of 209 and 557 amino acid residues . The two chains of the molecule are closely intertwined forming a pyramidal structure creating a cone-shaped dip at the bottom forming the active site. An N-terminal (serine) nucleophile acts as the catalytic residue which forms a four layer a+b structure with two anti-parallel b-sheets. The proposed mechanism of penicillin catalysis involves the nucleophilic attack of the terminal serine on the terminal a-amino group adjacent to each other. The residues (Arg A145, Phe A146, and Phe B71) are flexible and can change formation to respond to the presence of a ligand. Movements between the helical and coil conformations among these residues, allows the substrate access to the active site.
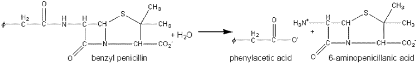
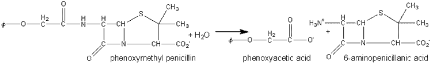
The intertwining of the two chains forms a cone shaped dip at the bottom forming the . This cone shape can be visualized better with the spin on. Once spinning, at a 180 degree rotation, you can see that the active site is actually buried deep within the enzyme or in the "cone".
The terminal serine, , plays a large role in the catalytic mechanism by nucleophilically attacking an adjacent amino group.
The following residues effect the conformation of the active site allowing the substrate access or restriction.