Home | Introduction | DNA Binding | DNA Separation | DNA Unwinding | Conclusion | References
When the ssDNA binds to the helix, the nucleotide binding site undergoes a very small conformation that moves a water molecule and an arginine residue into a position where the water can hydrolize the NTP to NDP by hydrophilic attack. The cleaved phosphate is then detected by nearby residues through hydrogen bonding interactions. When the free phosphate is detected, another conformational change occurs in the ssDNA binding site. The binding site returns to it’s original disordered state and DNA binding affinity plummets. The ssDNA and the NDP are both released.
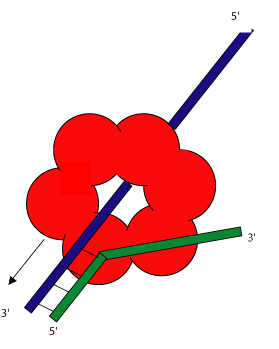
The ring shaped helicases of various organisms bind to double stranded DNA in very similar fashion. At a replication origin, the helicase either wraps itself around a single strand of the double stranded DNA, or the 5' end of a nicked strand is threaded through the central channel. The mechanism for this activity is as yet unknown. The complementary strand is then diverted around the outside of the ring. This is shown in the illustration to the left.
Each monomer of the hexameric ring contains one ssDNA binding site and one dsDNA binding site. The ssDNA is bound near the interface between pairs of adjacent monomers. The enzymatic monomers contain a mononucleotide-binding fold that binds dTTP, dATP, or dTDP.
This new helix contains two consecutive residues(arg487, gly488) that form the strongest bonds to the ssDNA and aligns it for more advantageous binding by the rest of the residues of the helix. The helicase, being a dimer, would then bind the ssDNA with two of the analagous helices. (This is a view of an E. coli Rep helicase binding to ssDNA.)