|
Glyceraldehyde-3-phosphate dehydrogenase (GADPH) is an important enzyme in glycolysis and gluconeogenesis, whose cycles occur in the cytoplasm. GADPH is responsible for catalyzing the reversible conversion of glyceraldehyde 3-phosphate (GAP) and inorganic phosphate into 1,3-bisphosphoglycerate (1,3-BPG) in a three-step reaction (12). Figure 1 (15) depicts this reversible reaction. The first and second step of the reaction involves the oxidation of GAP to a thiohemiacetal intermediate. The third step of the reaction phosphorylates the thiohemiacetal intermediate, which produces 1,3-BPG. During the catalyzation of GAP to 1,3-BPG, NADH is produced along with a proton, which is oxidized from GAP (15). GADPH is dependent on NAD+ cofactor, which acts as an electron acceptor, and inorganic phosphate (11). GADPH also contains two sulfate molecules per subunit. These sulfates represent the substrate phosphate (Ps) and inorganic phosphate (Pi), which are involved in catalysis (16).
Figure 1: The reaction mechanism catalyzed by GAPDH
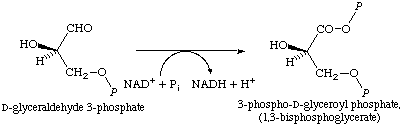
GADPH is a tetramer with a total molecular weight of 145 kD. All four subunits: O, Q, R, and P are independent of eachother, however each subunit contains the coenzyme NAD+ (11). Each subunit also contains 331 residues with molecular weights of 35.9 kD. The structure of GADPH, O and P subunits are composed of nine alpha helices and seventeen beta sheets. Whereas the Q subunit is composed of eight alpha helices and seventeen beta sheets, and the R subunit is composed of ten alpha helices and nineteen beta sheets. In all four subunits there are both parallel and anti-parallel beta sheets; that are flanked by a layer of alpha helices, which is characteristic of the Rossman fold (18). The standard free-energy change for the conversion of GAP to 1,3-BPG is 6.3 kJ/mol or 1.5 kcal/mol (15). Coupling its reaction to the thiohemiacetal intermediate lowers this reaction’s standard free energy.
GADPH is composed of two folding domains. The first domain contains the residues 0-148, which are involved in NAD+ binding. This domain is composed of a beta-alpha-beta pattern with a central beta sheet covered on both sides of the alpha helices. Whereas, the second domain contains the side chain residues 149-333, which are implicated in catalysis. The second domain is composed of extensive anti-parallel beta sheet regions. This domain also contains an S-shaped loop of polypeptide residues 178-201, which are in contact with NAD+ on the R axis. The S-loop also interacts with several amino acids across the P axis. The S-loop forms the core of the four subunits, and most of the residues are internal and essential for the interaction across the R axis (3).
The Q-axis, R-axis, and P-axis make up the three orthogonal twofold symmetry axes of GADPH, which relate the four subunits together as seen in Figure 2 (3). The P-axis interactions involve highly conserved residues from the anti-parallel sheets in the active site, and are produced by numerous hydrophilic and hydrophobic interactions. There are a total of twenty-six intersubunit hydrogen bonds; including seven salt-bridges and two bonds between main chain atoms. Interactions between R-axis related subunits involve the residues of the S-loop, which is also in contact with NAD+ and with the P-axis. The Q-axis contains few residues. However, Arg 281 from one subunit, and Arg 281 from the Q-axis related subunit form a double salt bridge with Gln 201 from both the R-axis and P-axis related subunits. These interactions bring the S-loop from two subunits into contact with the other two subunits (3).
Figure 2: Folding Diagram of one subunit of GADPH
GADPH is an acidic protein. On the surface of the active site there is an excess of negative charge. However, at the entrance of the active site pocket residues Lys 191, 211, and Arg 231 give an excess positive charge. The side chains of the S-loop, and coenzyme-binding domain contain an evenly distributed negative and positive charge (11).
The mechanism that is catalyzed by GADPH involves GAP reacting with GADPH to form a hemithioacetal intermediate (3). This intermediate involves two essential residues Cys 149 and His 176 (10). Cys 149 is located at the N-terminus of the alpha helix containing Ser 153. The Ser-Ser hydrogen bond links this helix to a beta strand, and provides stabilization in the active site (16). His 176 acts as a chemical activator by enhancing the reactivity of the thiol group of Cys 149 via the formation of an ion pair with the imidazolium ring of His 176. Hydrogen bonding between the protonated imidazole N6 of His 176 and the carbonyl oxygen of GAP, also stabilizes the hemithioacetal intermediate, and favors a nucleophilic attack by the thiol group of Cys 149, by lowering its pKa. His 176 also plays the role of a base catalyst, facilitating the hydride transfer from the hemithioacetal intermediate toward the C4 position of the nicotinamide ring of the NAD+ coenzyme during the oxidoreduction step (10). This hydride transfer results in the conversion of the hemithioacetal intermediate into a thioester intermediate and NAD+ into NADH (3).
After NADH dissociates from the active site, a second NAD+ molecule binds in its place. This action accelerates phosphorolysis and promotes specificity for phosphate instead of water as the acyl acceptor. The attacking phosphate in the transition state can be stabilized by hydrogen bonds with Ser 148, and Thr 150; the amide nitrogen’s of Cys 149, and Thr 150; and the C2 hydroxyl of the substrate (3). GADPH contains two sulfate molecules per subunit. These sulfates represent the two-anion binding sites: substrate (Ps) and inorganic (Pi) phosphates, which are both involved in catalysis. The Ps site is fully occupied by a sulfate ion Sul 388, which forms hydrogen bonds with Arg 231 and Thr 179. It is also hydrogen bonded to the 2’ hydroxyl of the nicotinamide ribose of NAD+, which is essential in the formation of Ps site. The inorganic phosphate is located in the Pi site, which is partially occupied by sulfate ion Sul 339, which is hydrogen bonded to the side chains of Ser 148, Thr 208, to the main chain of Gly 209, and to two water molecules, which are hydrogen bonded to Arg 195. The network of hydrogen bonds involving Ser 148 and Thr 208 forces the hydroxyl groups of these residues to act as proton donors in interactions with an anion in the Pi site. The Pi site also plays a role in binding C3 phosphate of substrate GAP and the hemithioacetal intermediate (11). The final step catalyzed by GADPH is the acyl-enzyme being phosphorylated to give the product 1,3-BPG (3).
|
|
|
|
| GADPH is composed of four independent subunits. Each chain is colored a different color and contains NAD, which is colored by atom type.
By clicking on the choices under "Color By" GADPH can be colored by it's secondary structure, group, or domain.
The domain option colors the first domain of GADPH blue. This domain contains the residues 1-148, which are involved in NAD+ binding. This domain is composed of a beta-alpha-beta pattern with a central beta sheet covered on both sides of the alpha helices. Whereas, the second domain, which is colored orange contains the side chain residues 149-333, which are implicated in catalysis. The second domain is composed of extensive anti-parallel beta sheet regions. This domain also contains an S-shaped loop of polypeptide residues 178-201, which are in contact with NAD+ on the R axis. The S-loop is colored green. The S-loop forms the core of the four subunits, and most of the residues are internal and essential for the interaction across the R axis (3).
|
|
| "NAD Interactions" contain the cofactor NAD and the residues it interacts with. NAD+ molecules are very similar in all four subunits. Therefore only one subunit will be shown. The adenine and nicotinamide rings are roughly perpendicular to the planes of the neighboring riboses (11). Residues Leu 187 and Pro 188 from the R-axis related subunit are in van der Waals contact with the adenosine ribose (3). NAD+ is situated in a cleft formed by the coenzyme binding domain, the S-loop, and another S-loop from the R-axis related subunit. The nicotinamide group resides deep in the active site pocket. The active site of GADPH is located at the bottom of a broad pocket made by the S-loop,which is colored green, and the two principal domains. At the nicotinamide end of NAD+ in the interior of the tetramer, the cleft opens to the spacious active site pocket, which is accessible by substrates. There are eight hydrogen bonds that are evenly distributed over structural elements of the coenzyme between NAD+ and the protein as seen in the below JMol image under "NAD Interactions". The pyridine ring of the nicotinamide takes part in hydrophobic interactions with the residues Ile 11 and Tyr 317 side chains,which are colored blue. The adenine ring lies in the pocket between the residue side chains of Leu 33 and Thr 96,which are colored purple (11). |
|
|
|