| HOME | INTRODUCTION | STRUCTURE | DNA BINDING | CLEAVAGE | REFERENCES |
DNA Binding
What Kinds of Interactions does a Restriction Endonuclease Have with DNA?
Restriction Endonucleases can non-specifically bind to DNA. This involves contacts to the phosphodiester backbone of DNA, not the bases. Once this occurs, the floor of the DNA binding site becomes more ordered, making it more compact. The catalytic site is still far away from the backbone, which allows the protein to diffuse or slide along the backbone, scanning for the restriction site. These vary from enzyme to enzyme: they are usually four to eight bases long and are palindromic. Different restriction enzymes that recognize the same restriction site are called isochizomers. The enzyme can scan up to a million basepairs in one binding event!
 |
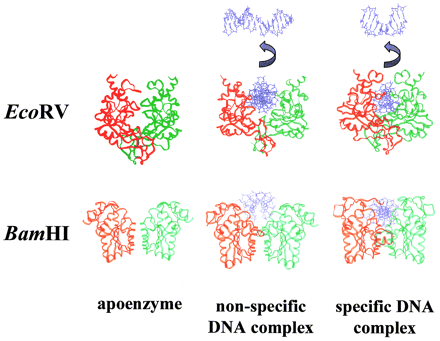
Here are drawings of three different structures of EcoRV and BamHI. The apoenzyme is what the free enzyme looks like. The non-specific complex is the enzyme bound to DNA at a place other than the restriction site. Note how it is more compact than the apoenzyme. The specific complex is the enzyme bound to the restriction site, and is even more compact than the non-specific comples. This serves to bring the catalytic site next to the backbone in order to cleave it. Also shown is the degree of DNA bending that occurs when EcoRV binds specifically (no other enzymes are known to bend DNA like this). It serves to better position the DNA for cleavage.
A metal ion is also required for most enzymes to specifically bind to the restriction site and for cleavage. EcoRV prefers magnesium or calcium, and some others prefer manganese.
Pictured below left is EcoRV apoenzyme. It is very loosely ordered compared to the specific complex on the right.
| Please push the buttons in order given. |