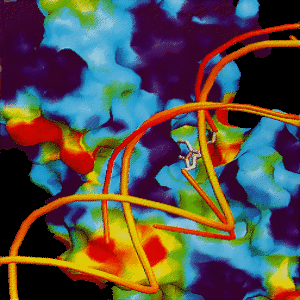
The DNA abasic site is shown in white and placed near the F-G beta-hairpin (red tube).

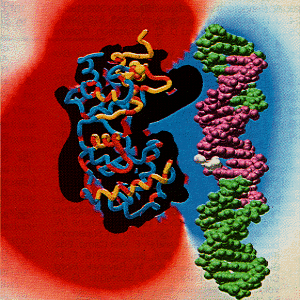
There are two novel DNA binding motifs consisting of a helix-hairpin-helix (HhH) and a [4Fe-4S] cluster loop (FCL). The FCL lies at the other end of the interdomain groove from the HhH. There are positive charges that are conserved on Lys 191, Arg 193, and Arg 190 in the surface exposed loop which implicates their side chains and the [4Fe-4S] loop region in potential DNA binding. Both binding motifs are dependent on the metal cluster for organizing the structure of the DNA binding loop (11).
Click here to see the HhH and FCL binding motifs. [13]
The DNA could be positioned against the positively charged electrostatic face of the protein and in both motifs, including the active site residue Lys 120. The DNA is aligned so that the side chain of Arg 190 could contact the major groove of DNA and Lys 191 and Arg 193 could contact the backbone of the DNA. The backbone of DNA fits into the interdomain groove of the
endonuclease III Lys 120 and solvent filled pocket. The DNA is bound in such a way as to allow an extrahelical base (11).
In endonuclease III there is a thymine glycol binding site, which is associated with the recognition of damaged DNA, adjacent to the main chain of the F-G loop. The F-G loop is a seven residue beta-hairpin that includes residues 113-119. The thymine glycol binding displaces two bound water molecules without significant conformational changes (6).
The F-G beta-hairpin loop causes unusual exposure of the main chain residues 112 to 118, which would allow close contact to DNA. The F-G beta-hairpin appears to be involved in substrate recognition and is positioned to provide a flexible reading head to accommodate different types of damaged DNA substrates (6).
There are two types of endonuclease III and DNA binding. One type is nonspecific electrostatic binding to DNA and the other is specific binding. In nonspecific binding the endonuclease III protein's positive charge follows the contours of B-DNA. The specific bond has a higher affinity of about 300 times that of nonspecific bonding and binds to damaged DNA by the flexible beta-hairpin region (6).
The DNA is aligned to endonuclease III at an angle of 20 degrees relative to the long axis of the enzyme, alpha E and alpha H would lie across the DNA minor groove. The F-G beta-hairpin is in the major groove (6). The endonuclease III enzyme binds along the DNA backbone, or binds to the minor groove, protecting one strand more that the other. Endonuclease III binds more tightly to the damaged strand where protection was observed (9).
There is a small binding site of about five to seven base pairs for specific binding to AP sites (9). There are approximately 12 base pairs bound to endonuclease III for non-specific interactions. Endonuclease III has a different confirmation for the two types of binding, which correlates with the five to seven base pair binding site for specific binding and the 12 base pair binding site for the nonspecific binding site (12).
Below is a table of models for DNA and endonuclease III binding.
The following pictures were obtained from reference [14].
 |
Association of the sequence-conserved face with a long stripe of positive electrostatic potential.
The DNA abasic site is shown in white and placed near the F-G beta-hairpin (red tube). |
 |
Accommodation of a flipped out base within the endonuclease III active site pocket. |
 |
Endonuclease III interdomain groove, electrostatic surface, sequence conservation and DNA binding model. |