The avidin tetramer has almost exact 222 molecular symmetry; the three possible dimers display quite distinct packing interfaces. The streptavidin protomer is organized as an 8-stranded beta-barrel. Pairs of the barrels bind together to form symmetric dimers, pairs of which in turn interdigitate with their dyad axes coincident to form the naturally-occurring tetramer. Each protomer is organized in an eight-stranded antiparallel orthogonal beta-barrel, with extended loop regions. The avidin binding site within each promoter is located in a deep pocket, at the center of the barrel, displaying both hydrophobic and polar residues for recognition of the tightly bound vitamin. Two Trp residues, Trp70 and Trp97, and Phe79 are in close contact with biotin. Moreover, the binding pocket is partly closed in its outer rim by residue Trp110 of a neighboring subunit. Once bound, biotin is almost completely buried in the protein core, with the exception of the valeryl side-chain carboxylate group which is exposed to solvent, hydrogen bonds to residues Ala39, Thr40 and Ser75, and triggers the formation of a network of hydrogen bonded water molecules.
Double-Click on the MPEG movie below to see various conformations of biotin binding to
streptavidin based on varying energy levels (kcal/mol).
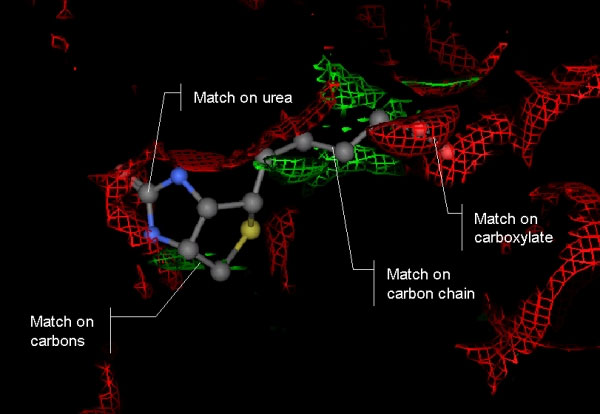
Shown here is a detailed schematic of how biotin binds with certain residues (named above) and the component parts of biotin that are "matched" for the Streptavidin protein-bound conformation.