Human Lysozyme
|
Introduction Human lysozyme was
discovered more than seventy years ago in 1922 by Alexander Fleming.
Alexander Fleming was a bacteriologist in |
. |
|
|
|
|
|
|
|
|
Reaction Mechanism Before getting into the reaction mechanism of lysozyme it important to review the structure of the ligand that lysozyme
cleaves. Like pac man eating little white
dots, lysozyme likes to
much (catalyze) the reaction that breaks a specific polysaccharide
apart. A polysaccharide is nothing more than the linking together of
sugar residues to form a chain. The polysaccharide of lysozyme reaction is made up of N-acetylglucosamine
(NAG) and N-acetylmuramic acid ( |
|
|
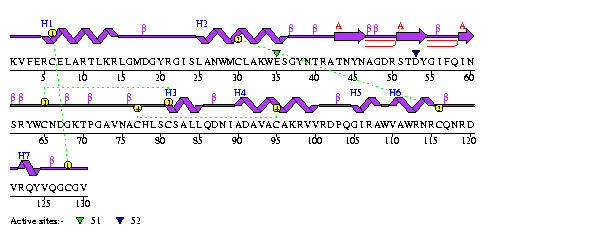
Structure: Overview Human lysozyme (HL) is
made up of 130 amino acids and has a macromolecular weight of 14,691daltons,
calculated by electrospray ionization mass
spectrometry.[Booth et al., 1997] It is a single strand enzyme with no subunits.
The secondary structures of HL can be broken down into seven helices and one
beta sheet consisting of three strands. |
|
|
|