 move on to page 5
move on to page 5 Sac7d Binds to multiple sequences of DNA.
| Sac7d has been produced recombinantly in E.coli and cocrystalized with 8-mers of heteroduplex DNA. The main chain folds into a nearly complete beta-barell of the OB topology, giving this protein its unique teritiary structure.
Sac7d has an overall secondary composition of 2 beta sheets and an alpha helix. Notice that both beta sheets are made of strands stabalized through hydrogen bonding side chains |
The supra secondary structure begins with a double pleated sheet, prior to turning through a small beta hairpin region connecting to a triple-pleated beta sheet. Push this button to highlight the two beta sheets. Notice that they lie on roughly perpindicular planes to one another. Pushing this button will select the helical residues and color them blue. Push this button to toggle spin mode on and off. Sac7d has been found to have robust thermodynamic properties. The teritary structure remains in tac, properly folded, up to temperatures equal to or exceeding 363 K. Because of this researchers wanted to see if sac7d can bind to DNA. Several nucleotide sequences (16nt palindromes or quasi palindromes) have been sequenced and found to bind to sac7d. Push this button to select the nucleic acid, colored according to base. |
|||||||||||
| RIGHT: Crystal structure of Sac7d model PDB 1sap, complexed with DNA sequence:
101 G 116 C |
||||||||||||
| RIGHT: Crystal structure of Sac7d model PDB code 1ca6 complexed the DNA nucleotide sequence:
101 G 116 C |
||||||||||||
| UP: Press this button to view Sac7d Getting dizzy? Hit this button. |
||||||||||||
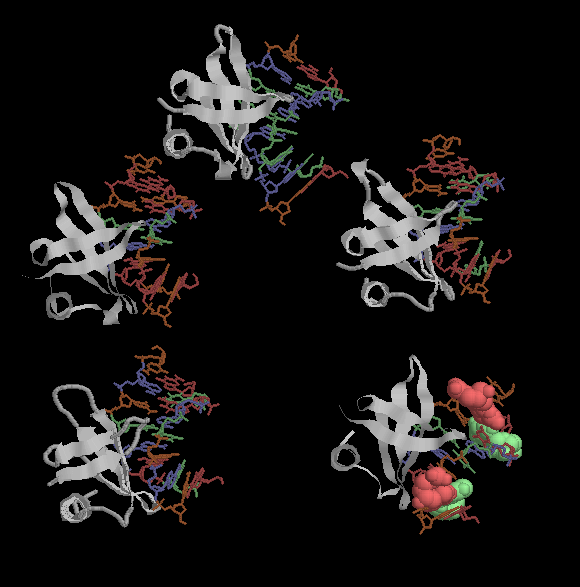
| Above: Clockwise starting from the bottom left; 1azp,1azq,1c8c,1ca5, and 1ca6. All of these proteins have slightly different nucleic acids and amino acid sequence, but retain a well conserved general form. 1ca6 has two G-T mismatch base pairs, here they are modeled in spacefill. sac7d is believed to preferentially intercalate at the less energetically stable T-G base pairs. | ||