Active Site
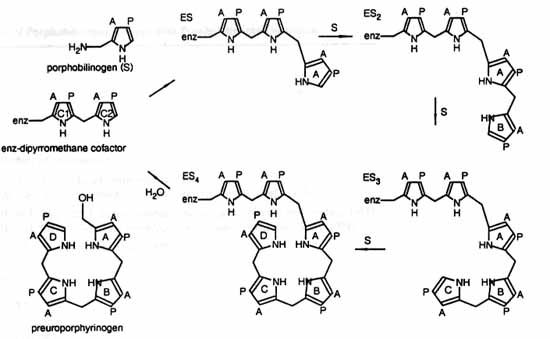
The mechanism of action and the active site of porphobilinogen
deaminase has been studied extensively. The mechanism of action
of porphobilinogen deaminase starts with deamination of porphobilinogen
to methylene pyrrolinene, which is subject to nucleophilic attack
by the carbon at the free alpha position of the terminal pyrrole(either
ring two of the DPM cofactor or any of the other terminal pyrroles
during chain elongation). Steric hindrance prevents chain elongation
from proceeding beyond a "hexapyrrole" so the product, preuroporphyrinogen,
is subsequently released by hydrolysis while the dipyrromethane
cofactor stays intact(see Scheme 1). The cofactor not only acts
as a reaction primer but it also limits the product to the length
desired(4,5,8,9).
Site-directed mutagenesis studies have established a number of
invariant residues required for full activity of the enzyme including
aspartate-84 and arginines 11, 131, 132, 149, 155, 176, and 232(2,3,4,8).
Cysteine-242, since the dipyrromethane cofactor is covalently
attached to it, is also essential for enzyme activity(7). A deep
cleft,
Scheme 1: Overall reaction(image from (8)p184)
Back to Main Page
Go to Conclusion Page

