Alkaline Phosphatase Continued
(pdb id 1alk*)
1alk was used because it was the enzyme not complexed with vanadate.
E. coli utilizes this (alkaline phosphatase) hydrolase as the method of attaining phosphate for use in the building of other important biological molecules like DNA, RNA, ATP, cAMP etc. The proposed catalytic mechanism by Kim and Wyckoff is as follows:
Scheme 2
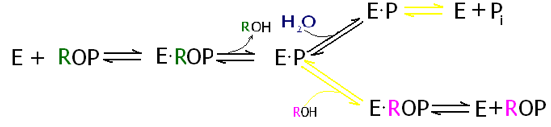
In this mechanism the ROP represents a phosphatemonoester. The enzyme (E) binds the ester, indicated by a “dot” to form the enzyme substrate complex. From here we can take either the top or bottom path. In the top path the OH portion of a water molecule breaks the phosphorous oxygen bond of the ester, and adds to the phosphorous forming inorganic phosphate (Pi), and the remaining proton that popped off of the water molecule is added to the remaining oxygen forming and alcohol. The bottom path is the same as the top, except in this case an alcohol is used instead of water. Note in the model below the Zinc cofactors (colored brown) which are essential for catalysis as well as the Magnesium ion (orange) that accompanies each pair. [4].
When the cofactors button is clicked I have also show Ser102 and Arg166. This was done because these two residues play the most critical role in catalysis. The ones that appear when "Key Sidechains" button is clicked are other residues which stabilize the substrate binding to the active site.
Mechanism of Rxn.
as Proposed by Holtz et al.
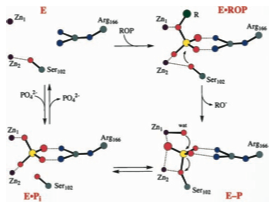
Alkaline Phosphate is found in the periplasm of the bacteria E. coli. From this location it can cleave the phosphate from any phosphate esters in its environment that make their way through the outer membrane. It can then be internalized and used in the cytoplasm. Its location in the periplasm also makes it one of the easier proteins to isolate because it can be removed without spilling the cytoplasm. and its overall durability makes it easy to purify. For these reasons it has been extensively studied. Alkaline Phosphatase is very non-specific; and will catalyze the above reaction in any phosphate ester provided that there is not much steric hindrance in the phosphate entering the shallow active site pocket. Upon clicking the second button above notice how exposed the active site is, located at the enzymes surface.