Introduction:
Lipoamide dehydrogenase, also called dihydrolipoyl dehydrogenase, is a flavin adenine dinucleotide (FAD) dependant enzyme that is a member of the pyridine nucleotide-disulfide oxidoreductases. Members of this group – lipoamide dehydrogenase, glutathione reductase, thioredoxin reductase, and mercuric reductase – are flavoproteins that contain a redox-active disulfide at each active site. All members function only as dimers and have a high degree of homology in their amino acid sequences; especially at the active site, the FAD-binding region, and the pyridine nucleotide binding region.[8]
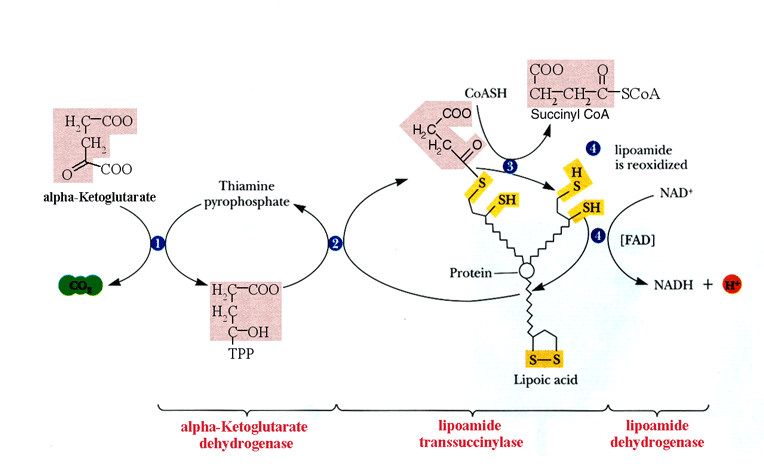
Lipoamide dehydrogenase (E3) is the terminal enzyme in both the pyruvate and a-ketoglutarate dehydrogenase complexes. In both of these complexes, E3 catalyzes the reaction:
Lip(SH)2 + NAD+ ===> LipS2 + NADH + H+.
This reaction oxidizes dihydrolipoamide (Lip(SH)2) that is the catalytic moiety of dihydrolipoyl transsuccinylase (E2), the second enzyme of the complex. As shown in figure 1, the a-ketoglutarate dehydrogenase complex removes a carbon dioxide molecule from a-ketoglutarate on the first enzyme, a-ketoglutarate dehydrogenase (E1). E2 then replaces this abstracted carbon with a CoA group producing succinyl CoA.[2] Finally, E3 accepts the elections gained by E2 and passes them on to the ultimate acceptor – NAD+ - oxidizing the lipoic acid on E2.
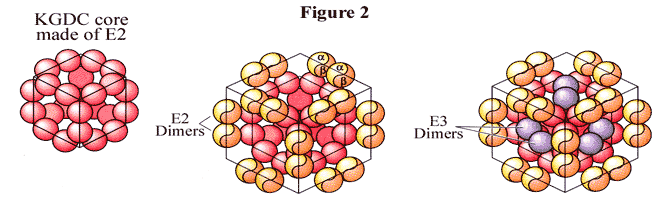
The a-ketoglutarate dehydrogenase complex (KGDC) is a cube shaped complex that incorporates E1, E2, and E3. In E. coli, 24 polypeptide chains of E2 form an octahedrally symmetric core which contains binding sites on its surface for E1 and E3 (figure 2); in mammals, this core is composed of 60 copies of the E2 subunit.[1] Note that E1 and E3 are not bond together but are bound to the complex by E2 alone. 12 polypeptide chains, present as six dimers, bind to the KGDC. The complex of E. coli can bind up to 12 more dimers of E3 beside the six already present; however, maximal activity doesn’t increase when surplus E3 is present. Nonetheless, all the surplus sites are functional; maximal activity of KGDC is not increased by surplus E3 because the reactions catalyzed by E3 are not rate-determining in the overall sequence of reactions at standard state conditions. This reaction can be made rate-limiting by adding an inhibitor, such as NADH, or by using low [NAD]; thus, under such conditions, excess E3 can increase the activity of KGDC. [11]