|
|
|
Biological Functions of Fumarate Reductase/Succinate Dehydrogenase
|
|
|
|
|
|
|
|
This page endeavors to relate some of the structural features of FRD and SDH to their functional roles.
|
|
|
|
Figure 1 |
|
|
|
 |
|
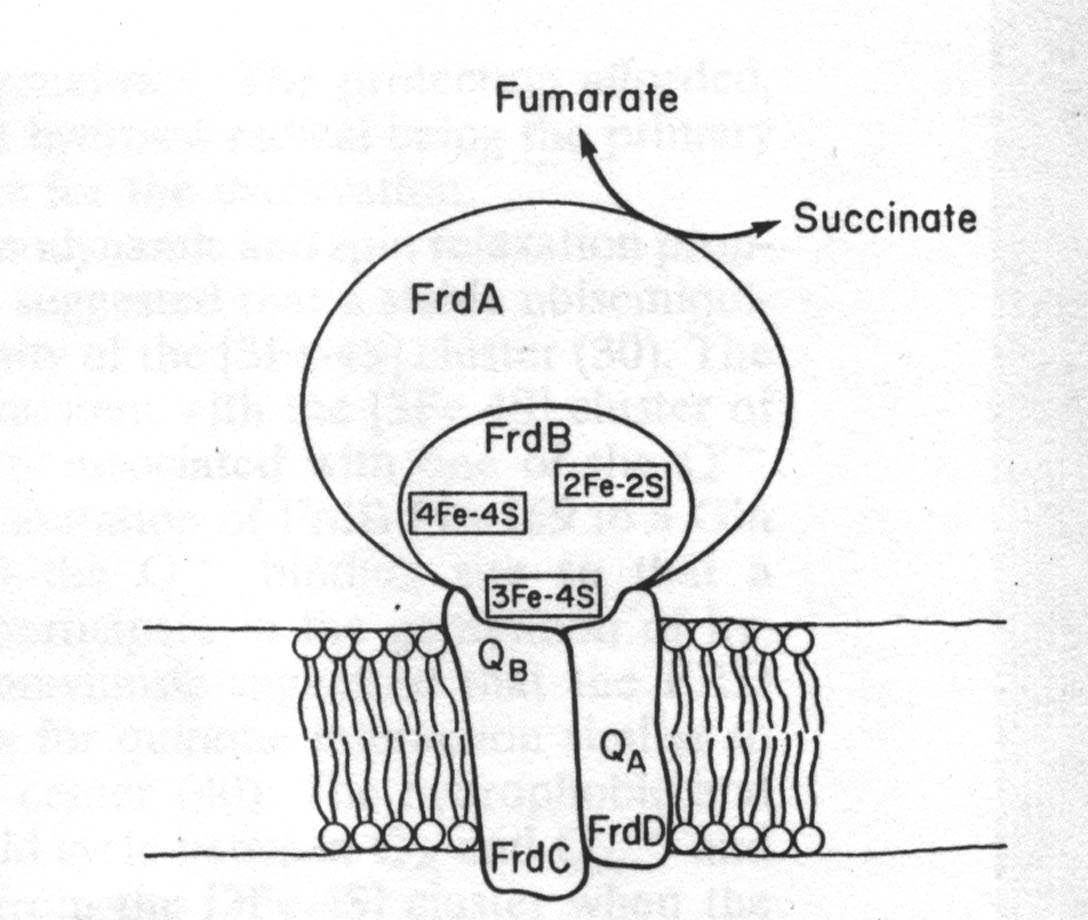
| Topographical model of native FRD showing the close spatial relationship between the[3Fe-4S] cluster in FrdB and the quinone (Q) binding sites in the hydrophobic FrdC and FrdD peptides |
|
|
|
As shown in the previous structure page, each FRD monomer consists of four domains. The first and largest domain, FrdA, makes up part of the catalytic domain of the enzyme. Within the amino acid sequence of this domain there exist three very conserved sequences. They include histidine 232, cysteine 247, and arginine 248. Many studies concerning the functions of these a.a's and their role in active site function were conducted in the late '80s and early '90s. It has been shown that loss of both fumarate reductase and succino-oxidase activities occured following site-directed mutagenesis of His-232 and Arg-248. The effects of removing histidine-232 of FrdA are consistent with its proposed role as a general acid-base catalyst. Cysteine-247 of FrdA was not shown to be essential for catalytic activity or the tight-binding of oxalacetate. This is a strange finding considering that Cys-247 is the only conserved cysteine in the flavoprotein subunit of fumarate reductase. As mentioned, these studies were performed on E. coli a little over a decade ago. Earlier this year (2000), some observations were made which contradict some of the previous findings. These studies were performed on Shewanella and Wolinella. Using different species causes a difference in the amino acid designation numbers, but the active site is held constant. It was found that the histidine residues are involved in Michaelis complex formation and are not essential catalytic residues. It has thus been determined that the only plausible active site would be the conserved arginine residue. This reaction mechanism for fumarate reduction involving arginine is found below in Figure 2. |
|
|
|
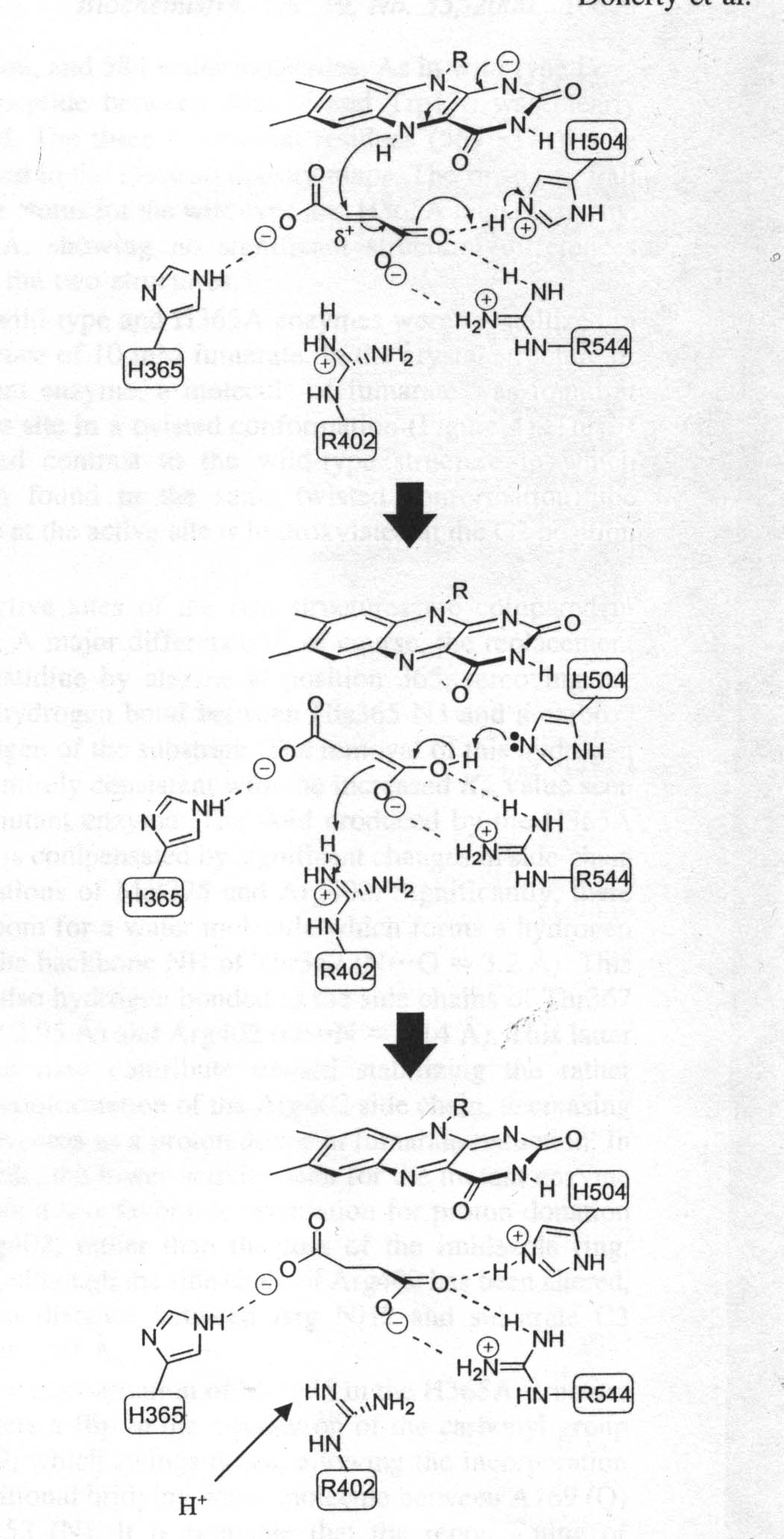 |
Figure 2
Reaction Mechanism for fumarate reduction. Catalysis is initiated by the twisting out of plane of the C1 carboxylate group (on the left) of fumarate. The substrate is polarized by interactions with charged residues facilitating hydride transfer from N5 of reduced FAD to the substrate C2. Arg is immediately reprotonated via a proton pathway involving Arg381 and Glu378.
|
|
|
| The FdrB subunit, which is located directly below the FdrA subunit, is also part of the catalytic domain of the FRD enzyme. The afforementioned iron-sulfur clusters found in this FrdB subunit, center 1,[2Fe2S] (Em=-20mV), center 2, [4Fe-4S] (Em=-320mV) and center 3, [3Fe-4S] (Em=-70mV) are essential for interactions with quinones. From studies conducted in 1995 it was determined that the [3Fe-4S] cluster is intimately associated with one of the quinone binding sites found in fumarate reductase and succinate dehydrogenase. This contention came about after observations made from site-directed mutagenesis on Pro-159, which is conserved in all FRDs and SDHs identified thus far. It is situated next to Cys-158, which partially ligates the [3Fe-4S] cluster. Mutation to a Gln or His causes inactivation of the mutant FRD complex during aerobic incubation with succinate. This phenominom only occurs in the membrane-bound enzymes. This supports the theorizations that the [Fe-4S] cluster is intimately involved in the site(s) of interaction of FRD with quinones and may be part of one of the quinone binding sites. |
|
|
| Also present in FRD are the subunits FrdC and FrdD. They are both small, 15 and 13kDa, respectively, and consist of mostly hydrophobic peptides. They are essential for the electron interchange between the enzyme, menaquinol, and ubiquinone. They are also responsible for attachment of the catalytic subunits to the inner surface of the cytoplasmic membrane. This is due to the hydrophobicity of the subunits. These two membrane anchor subunits, FrdC and FrdD are devoid of prosthetic groups, except for the enzyme from Wolinella succinogenes. This FRD enzyme (used in the structure page) contains a single membrane subunit which is a diheme cytochrome b. In FrdC and FrdD subunits, menaquinone is presumed to be bound at two sites, termed QA and QB, as part of the FRD redox chain. Addition of succinate to the enzyme would cause reduction of the [3Fe-4D] cluster, which in turn would reduce QA to QB-. The hydrophobic and tightly bound QA site would cycle between QA and QA - and take an electron directly from the [3Fe-4S] cluster when the enzyme is reduced by succinate. The somewhat more hydrophilic QB site can cycle between QBH2,QB-, and QB. It would appear that the FrdB Pro-159-to-glutamine mutation perturbs either the QA or QB site, thus allowing generation of hydroxyl radical which subsequently incactivates the enzyme. |
|
 |
|
 |
|
|
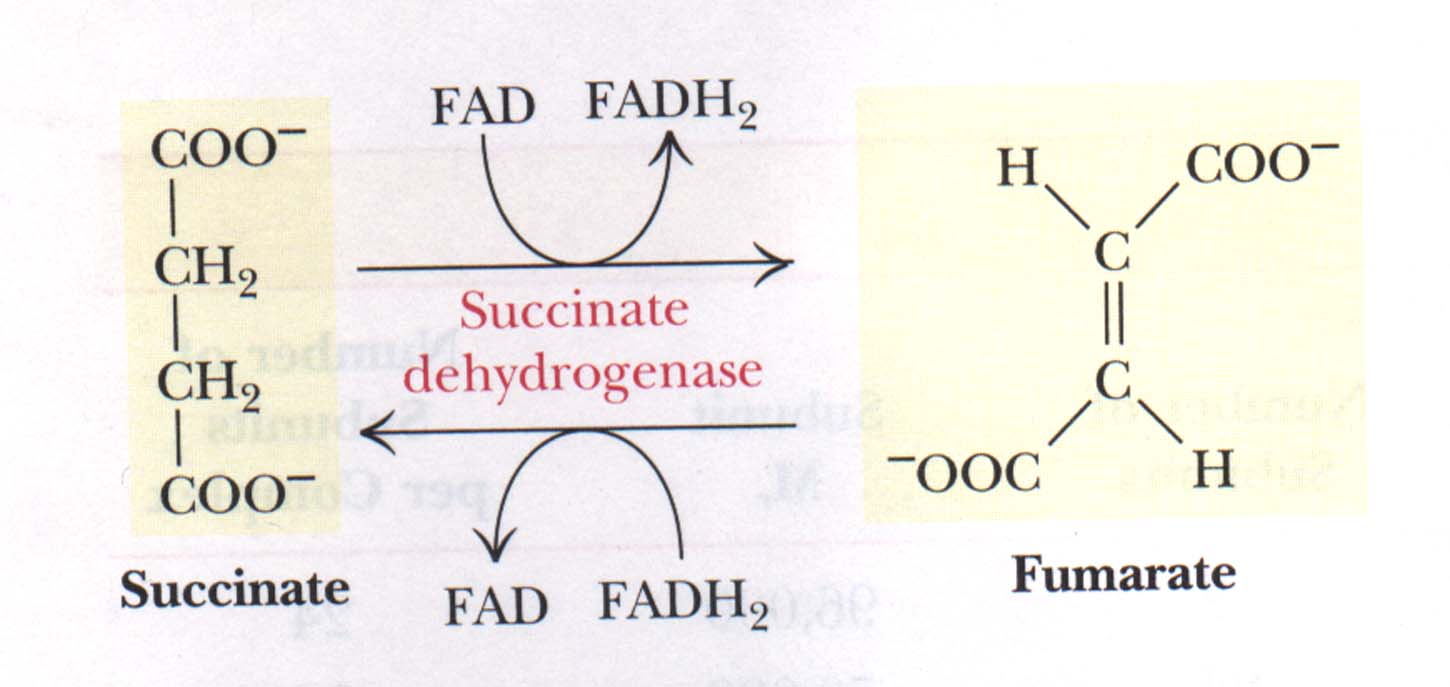
| Figure 3
The succinate dehydrogenase reaction. Oxidation of succinate occurs whith reduction of [FAD] and reoxidation of [FADH2] transfers electrons to coenzyme Q.
|
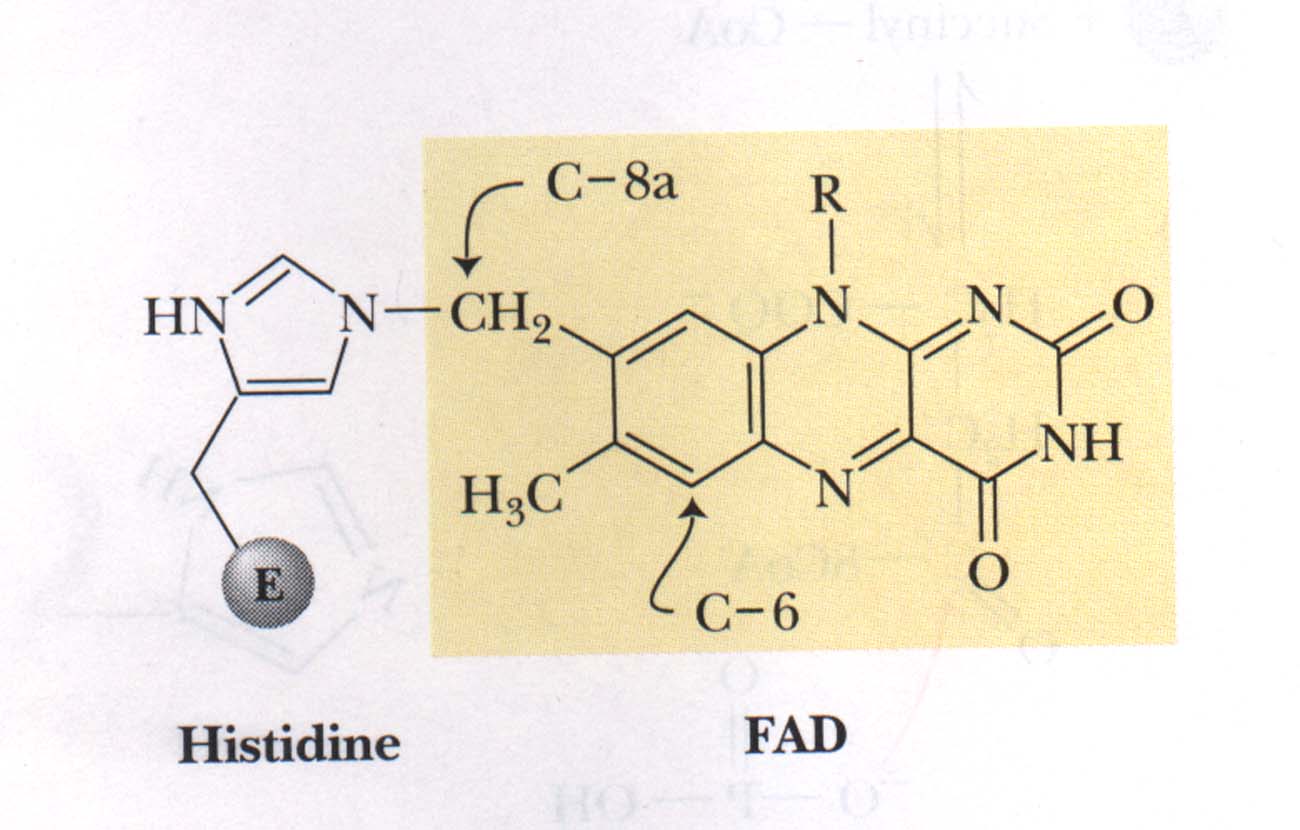
Figure 4
Shows the covalent bond between FAD and succinate dehydrogenase involving the C-8a methylene group of FAD and the N-3 of a histidine residue on the enzyme.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|



