From Escherichia Coli
(Push buttons in the order they come to see the full affect)
L-Aspartate is one of the allosteric effector molecules for PEPC that causes inhibition of the catalytic activity (Kai, 1999). The binding domain for the allosteric inhibitor consists of four amino acid residues, Lys-773, Arg-832, Arg-587, and Asn-881. All four of these residues are highly conserved in all PEPCs so the information stated just doesn't hold true for E. coli. Lys-773 and Arg-832 forms a salt-bridge with the carboxyl group in the side chain of aspartate. Arg-587 has 6 glycine residues surrounding it that provide a long flexible loop which is locates the allosteric site near the enzymes active site. The Arg-587 also forms a salt-bridge to the carboxyl group of aspartate, but unlike the Lys and Arg-832, Arg-587 is trapped by the aspartate. The Asn-881 forms hydrogen bonds with the aspartate.
(Click here to view the allosteric binding domain)
Lys-773= red; Arg-832=blue; Arg-587=green; Asn-881=purple
The active site is located at the C-terminus of the beta barel. Two highly conserved histidine residues are located in this area and are important for the catalytic activity. Histidine-138 and Histidine-579 can be seen in Fig. 4, from Fig. 6 by Kai, et. al., 1999, along with another important residue Arg-587. Arg-587 is involved in the allosteric binding of L-Aspartate. The interatomic distances between the alpha-carbon atoms of the histidines and the aspartate range from 17 angstroms to 18 angstroms (Kai et. al., 1999).
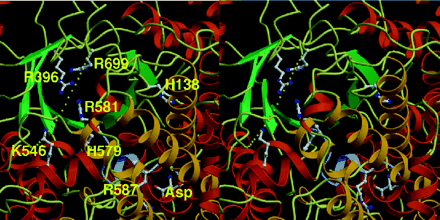
The residues involved in the catalytic active are shown in ball- and- stick representation. Can you identify the important residues His-138, His-579, and Arg-587? The missing loop is shown as dots. The beta sheets of the beta barel are shown in green, while the yellow and red regions are the alpha helices. The residues are all located at the C-termini. The missing loop is missing Lys-702 to Gly-708. It is here that the active site is located. Lys-702 and Arg-704 are highly conserved in all PEPCs identified. The missing loop, consisting of Gly-707 and Gly-708, trap essential functional residues with the help of Ala-701 or Ser-701. The link between the catalytic site and the allosteric binding domain is the glycine loop and the loop consisting Arg-587 and Arg-581. These two loops work together to catch substrate molecules at the active site with the their functional residues; plus they form lids protecting the reaction intermediates from outside water molecules (Kai et.al., 1999). The C-terminus of the beta barel contains 19 residues and 14 of these residues are hydrophobic making them essential for keeping water away from the reaction.
His-138=red; His-579=blue; Arg-587=green