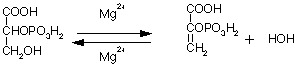
Enolase (2-phosphoglycerate hydrolase, E.C. 4.2.1.11) is a glycolytic enzyme that catalyzes the conversion of 2-phosphoglycerate (2-PGA) into phosphoenolpyruvate (PEP) by removing a water molecule. The enolase reaction is one of the final reactions in the glycolytic pathway, and follows the conversion of 3-phosphoglycerate into 2-phosphoglycerate by 3-phosphoglycerate mutase. Enolase also catalyzes its reverse reaction, the hydration of phosphoenolpyruvate into 2-phosphoglycerate, during gluconeogenesis.
Reaction 1: The reversible dehydration of 2-PGA into PEP
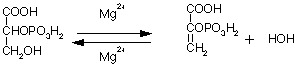
The free energy change of this reaction is fairly small, at about 1 kcal/mol under physiological conditions.3
Enolase reactions are reactions characterized by a proton (H+) shift from a carboxylate donor (from the enzyme) to a hydroxyl (~OH) group acceptor (from the substrate), forming water (H2O). The loss of an additional proton by the substrate results in the formation of an enol product.
Enolase is found in both eukaryotes and prokaryotes. The active site region and nearby secondary structure has been found to be highly conserved among all species studied.
In order for enolase to maintain its catalytic activity, divalent metal cations must be present. These metal ion cofactors essentially act as controls to activate or deactivate the enzyme. The "ideal" cofactor is Mg2+, which results in the highest activity level for the enzyme..6 However, several other divalent metal ions are also sufficient, including Zn2+, Mn2+, Co2+, and Ni2+. The first metal ion, the "conformational" cation, binds to the enzyme and causes a conformational change at the active site. This conformational change results in the ability of the substrate (2-PGA) to bind at the enolase active site. Once the substrate has been bound, a second, "catalytic" divalent metal ion is then needed to bind to the enzyme in order to enable the catalytic ability of enolase to become activated. Among the few metal ions that cannot invoke this activation are Ca2+ and Sm3+, both of which bind strongly to the enzyme at the conformational ion site, but fail to elicit sufficient conformational change to enable the enzyme to become activated and complete the dehydration reaction. Neither Ca2+ nor Sb3+ can alter the conformation of the enzyme enough to allow a second ion to bind near the active site, thus inhibiting activation by the catalytic metal cation. However, both of these ions bind to the enzyme more strongly than the activating cofactors, thus competing with the activating metals and acting as "deactivators" of the enzyme.
The metal ion binding sites in the three-dimensional structure of enolase are referred to as Site I (conformational) and Site II (catalytic) respectively. While the location of Site I in the structure has been known for some time, the exact location of site II in the enzyme has not yet been determined through crystallographic research and is an area of investigation.6
Following the enolase reaction in the glycolytic pathway is the conversion of PEP into pyruvate. This dephosphorylation is catalyzed by pyruvate kinase. To learn more about pyruvate kinase and other glycolyic enzymes, go to http://www.chem.uwec.edu/Chem406_F99/Pages/webpapers_F99.html.
|
Homepage |
|
Site |
|
Home Page |
Updated: Friday, December 10, 1999
By Christina Golner
Chem 406: Biophysical Chemistry